Illumina 's next-generation sequencer Genome Analyzer was first developed by Solexa , using its patented core technologies " DNA cluster" and " reversible terminator " for automated sample preparation and genome multi-million bases Parallel sequencing. Illumina company spent 600 million dollars in huge amounts of money in 2007 acquired Solexa, is to facilitate the commercialization of the Genome Analyzer. As a next-generation sequencing technology platform, Genome Analyzer has the advantages of high accuracy, high throughput, high sensitivity and low operating cost. It can simultaneously perform traditional genomics research (sequencing and annotation) and functional genomics (gene expression and regulation, Gene function, protein / nucleic acid interaction studies. With advanced Illumina high-throughput sequencing platforms , professional research teams, and personalized experimental solutions, Miki Bio enables you to create high-quality, high-throughput sequencing services.

experiment process
1. Prepare the library
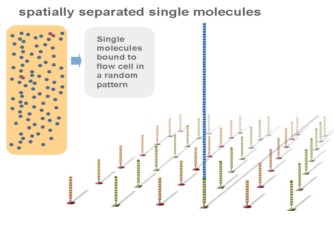
The genomic DNA is divided into small fragments of several hundred bases (or shorter) and an adapter is added at both ends .
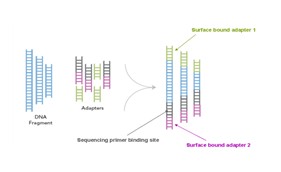
2 , bridge PCR to generate DNA clusters
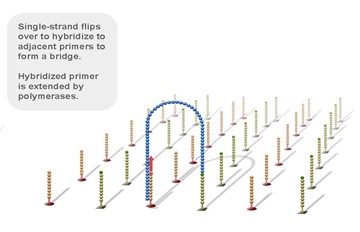
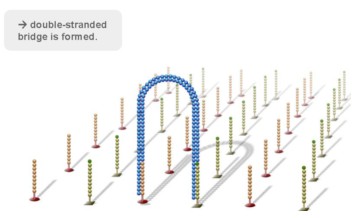
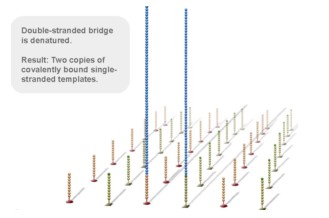
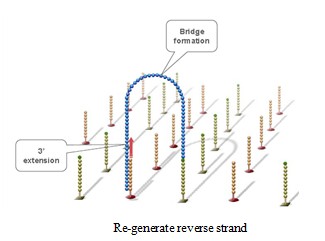
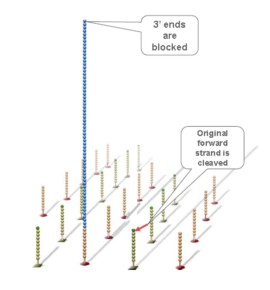
Using a patented chip, a single-stranded primer is attached to the surface, and the DNA fragment is single-stranded and then "fixed" to the chip by one end complementary to the primer base on the surface of the chip. The amplification of the primers causes the single-stranded DNA to become double-stranded, and the double-stranded strand becomes a single strand, one end of which is "fixed" on the chip, and the other end ( 5 ' or 3 ' ) is randomly complementary to another primer in the vicinity, and is also " fixed "live, to form a" bridge (bridge). " Repeated 30 rounds of amplification, each single molecule was amplified 1000 times, becoming a monoclonal " DNA cluster".
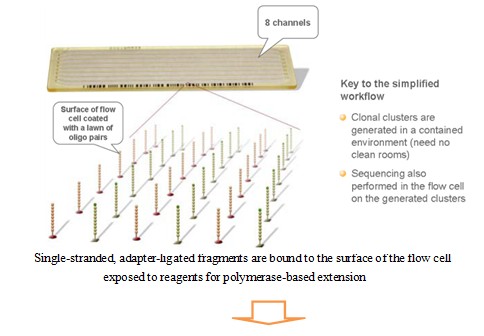
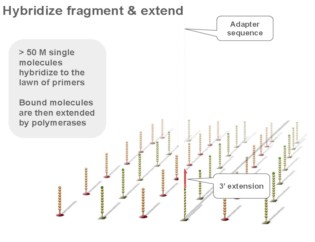
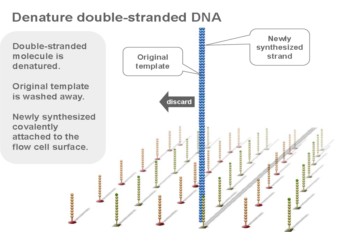
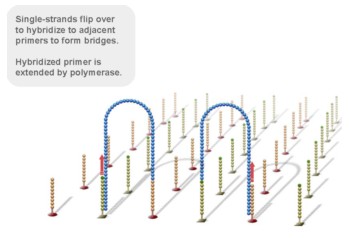
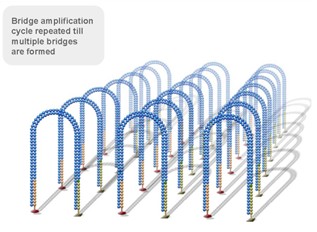
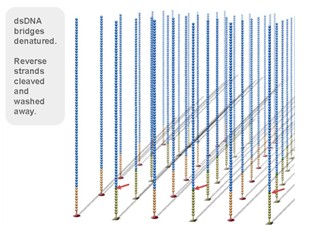
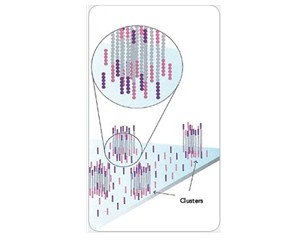
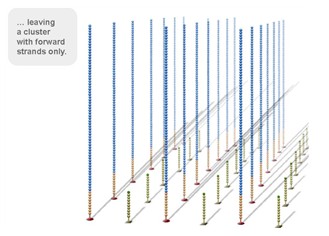
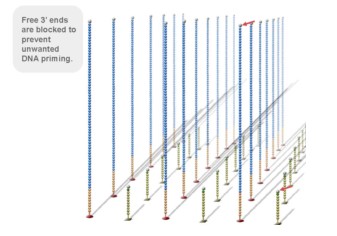
3, reversible chemical blocking sequencing technology
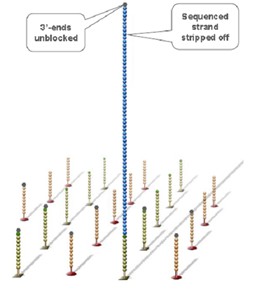
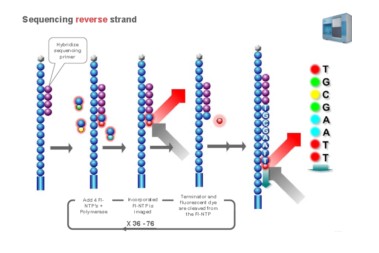
Using sequencing-by- synthesis (Sequencing b y s ynthesis) principle, added transformed with DNA polymerase and four kinds of fluorescently labeled dNTP. These nucleotides are " reversible terminators " because the 3' hydroxyl terminus carries a chemically cleavable moiety, which allows only a single base to be incorporated per cycle. After removing other excess dNTPs , the surface of the reaction plate was scanned with a laser to read the nucleotide species polymerized in the first round of each template sequence. These groups are then chemically cleaved to restore the 3' end viscosity and continue to polymerize the second nucleotide. This cycle until each template sequence is completely polymerized into a double strand. The sequence of each template DNA fragment is known by counting the fluorescence signals collected in each round .
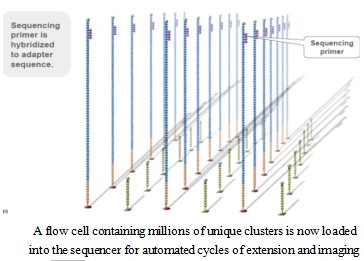
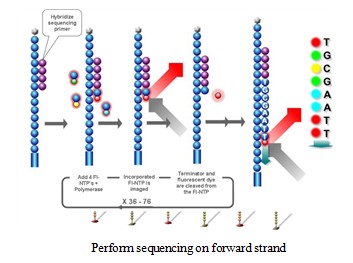
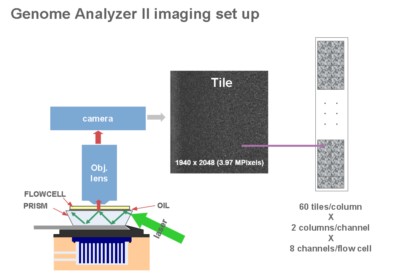
4, data analysis
The base is automatically read and the data is transferred to an automated analysis channel for secondary analysis.
Technical advantages
★ Innovative sequencing chemistry technology
The Genome Analyzer utilizes novel reversible fluorescently labeled terminators to detect single base incorporation during DNA strand extension. Since four reversible terminator dNTPs are present in each sequencing cycle, natural competition reduces the error of incorporation.
★ Scalable high throughput
The Genome Analyzer system can get over 20Gb of high quality filtered data after each paired end run . The scalability of this technology guarantees higher data density and output, allowing for more complex projects with less expense.
★ need less sample size
The Genome Analyzer system requires as little as 100 ng of sample and can be used in a limited number of samples, such as immunoprecipitation, microdissection, and more.
★ Simple, fast, and automated
The Genome Analyzer system provides the simplest and simplest workflow. Sample library preparation can be completed in a matter of hours, and high-accuracy data can be obtained in one week. The process of automatically generating DNA clusters controlled by independent software can be completed within 5 h ( 30 min manual operation). Automated processes do not require water-in-oil PCR , reducing the possibility of manual handling errors and contamination, without the need for robotic operations or cleanrooms. The rapid experimental process maximizes the capabilities of the Genome Analyzer , which reduces the time and expense of the project.
★ single or paired end support
The Genome Analyzer system supports single fragment or paired end libraries. The library construction process is simple, reducing the time required for sample separation and preparation. It takes 6 h to prepare a single fragment or paired-end library of genomic DNA , and only 3 h of manual manipulation .
Platform advantage
★ Flexible platform
For different applications, it is possible to choose whether to combine different read lengths with read sequencing techniques.
★ high quality sequencing data
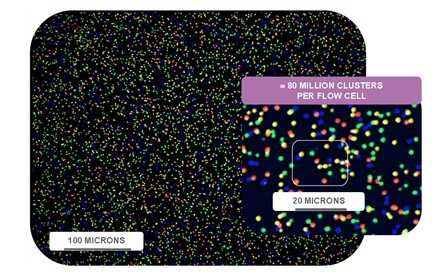
Accurate base identification of tens of billions to hundreds of billions of bases in each flow cell.
★ Wide range of applications
SNP and structural variation detection, ( de novo ) assembly, transcriptome sequencing, methylation detection.
★ Efficient sample preparation
High-speed automated workflows and low sample volume requirements generate data in less than a week.
Technical application
Ø Transcription analysis
l mRNA sequencing (RNA-Seq)
An mRNA sequencing experiment can quickly generate complete sequence information of complete poly-A -tailed RNA , and analyze gene expression, cSNPs , brand new transcription, brand new isomers, splice sites, allele-specific expression and rare Transcription.
l Small RNA deep sequencing
Small RNA sequencing is capable of measuring small RNA sequences of any size from different organisms . Simultaneous analysis of 4 million small RNAs in a single sample is the most widely used and most intensive small RNA detection method available. Small RNA sequencing records the digital frequency of the sequences in the library population, thus eliminating the background signal.
l Digital gene expression profiling
Digital gene expression profiling by high-throughput sequencing on behalf of each gene 21bp tag (tag) to quickly and comprehensively detect the expression of a gene in a specific tissue species under certain conditions. Because label sequencing eliminates the need to determine the total length of the expressed gene, it has fewer total reads and is more sensitive, allowing researchers to adjust the depth of coverage to a near-unrestricted range, completing rare transcriptional recognition and quantification.
Ø Gene regulation and control
Illumina ChIP-Seq integrates chromatin immunoprecipitation ( ChIP ) and massive parallel DNA sequencing technology to accurately and cost-effectively identify and protein-bound DNA sites. ChIP-Seq technology provides powerful insight into ChIP proteins and modifications. Technical Support.
The ChIP method utilizes a specific protein in an antibody to specifically enrich the cross-linked DNA- protein complex. The DNA linker and the specific protein-binding DNA are then spliced ​​to allow massive parallel sequencing. After selection according to the size of the fragment, the gene analyzer and Illumina sequencing technology are used to sequence the generated ChIP DNA fragment, and low resolution. different rates of chIP-chip technology, the time sequence for the accuracy of genome-wide association analysis of chIP-Seq.
ØMultiple sequencing
Illumina oligonucleotide to provide multiple sample preparation kit, multiplex sequencing primers and PhiX Control Kit, can be introduced marker sequence (tag) in the DNA fragment in order to achieve 96 different samples are sequenced in a flow channel simultaneously. Under the condition of ensuring low error rate and reading length, the scalability of the experiment is greatly improved. Multiple sequencing is more valuable, especially when studying a region or minigenome.
CE Certified 3 PLY Surgical Medical Face Mask, EO-Sterile / Non-Sterile:
These Surgical Masks Disposable Medical Grade are class I medical devices, and are intended for use by operating room personnel during surgical procedures to protect both the patient and the medical personnel from transfer of microorganisms, bodily fluid, and particulate material. Bacterial Filtration Efficiency ≥ 99%. Medical grade disposable face mask features tri-layer protection and breathable lightweight design. Surgical Mask Antiviral Filters out harmful substances, such as dust, bacteria*, air pollution, etc. Comfortable ear loops and form-fitting flexible nose bar.
Surgical Masks Disposable,Surgical Face Masks Medical,Surgical Mask Antiviral,Surgical Masks Disposable Medical Grade
Suzhou JaneE Medical Technology Co., Ltd. , https://www.janeemedical.com