Release date: 2018-05-17

This article is reproduced from the "Life Mystery", the original title: a new generation of diagnostic technology built on the basis of CRISPR technology.
The rapid and accurate diagnosis of infectious diseases is the key to improving the level of clinical diagnosis and treatment, as well as guiding the prevention and control of infectious diseases and public health work, and avoiding the spread of immunity. This is true for large modern medical centers and lack of medical care. The remote township hospitals are very important. The most ideal diagnostic method should be low-cost, high-accuracy, and fast, and should be easy to operate (no need for professional operation), and no need for professional equipment and infrastructure requirements (such as electricity). If you have this ideal for pathogenic viruses such as Ebola virus and Middle East respiratory syndrome coronavirus that occur in remote locations but cause widespread global spread. The detection technology can help medical staff to find patients at an early stage, isolate them, avoid the spread of immunity, and also give patients more opportunities for timely treatment.
Studies have found that prokaryotes such as bacteria and archaea have innate adaptive immunity, and the immune system is based on the recently popular gene editing technology CRISPR-Cas system. based on. The recent Science magazine has introduced the work of the Chen research team, the Myhrvold research team and the Gootenberg research team. They have developed a revolutionary new molecular diagnostics technology using CRISPR-Cas technology to discover Zika virus (ZIKV), Dengue virus (DENV) and human papillomavirus in human specimens ( Human papillomavirus, HPV). At the same time, they also used circulating DNA-free DNA of lung cancer patients as specimens to diagnose non-infectious pathogenic factors such as genetic mutations.
Prokaryotes store genetic information on a large number of infectious pathogens (such as phage, plasmids, and transposons) in the CRISPR site of their genome. These genetic information is the "antibody (adapted immune memory)" of the prokaryote itself. The Cas protein completes the adaptive immune response of prokaryotes through steps such as adaptation, transcription of crRNA, and interference. In the adaptation stage, exogenous genetic material is processed and selected by prokaryotic organisms, and finally integrated into the CRISPR site to lay a deep imprint of immune memory, ready for the future encounter with the source of infection. The precursor crRNA is a long-chain RNA precursor molecule. After it matures into a crRNA, it can direct the Cas protein to cleave the complementary strand of the foreign genetic material. This is the interference step, which ultimately degrades the foreign genetic material and eliminates the foreign infection source. . After researchers have thoroughly clarified this set of prokaryotic immune mechanisms, they have developed new molecular diagnostic techniques.
Chen's team found that when the CRISPR-Cas12a protein cleaves dsDNA in a sequence-specific manner, it also induces a strong non-specific ssDNA trans-cleavage effect. Based on this feature, they developed a new technique for rapid identification of HPV 16 and 18 oncogenic viruses using clinical routine sampling specimens. The dsDNA of the HPV virus was extracted from an anal swabs specimen, subjected to isothermal preamplification, and then amplified by recombinant polymerase amplification (RPA). This DNA amplification technology is a fast and no need for specialized instrumentation for DNA amplification. Next, they bind to the amplified HPV dsDNA through the Cas12a-crRNA complex, decompose it, and induce dsDNA trans-degradation reaction to activate the fluorescent reporter bound to the dsDNA molecule, fluoresce and complete the whole. Detection. The new diagnostic technology is called DNA endonuclease-targeted CRISPR trans reporter (DETECTR). This technology can be used to quickly and accurately detect HPV viruses of various subtypes. And because of its fast, accurate and convenient features, it is especially suitable for large-scale census work recommended by the World Health Organization.
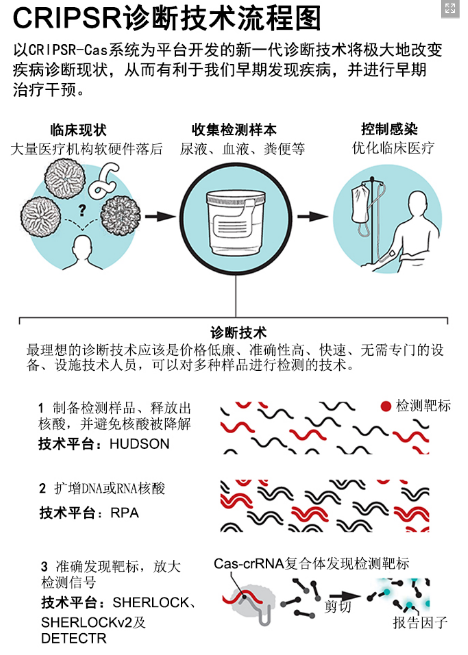
The detection method developed by Myhrvold et al. is to directly release the viral nucleic acid in the clinical test sample taken from the patient for detection, skipping the step of nucleic acid extraction in the routine detection work. This method is called heating unextracted diagnostic samples to obliterate nucleases (HUDSON). This technology can inactivate a large number of ribonucleases in human body fluids, and at the same time, it can rupture the capsid of the virus and release the nucleic acid molecules of the virus. Myhrvold et al. also used a specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) platform based on Cas13 protein. Similar to the DETECTR system, the SHERLOCK system also utilizes RPA and Cas13 proteins to induce (non-CRISPR) nucleic acid cleavage reactions.
In the SHERLOCK system, the RNA that binds to the fluorescent reporter factor is cleaved to release a fluorescent signal, which is then amplified by the enzyme, thereby greatly increasing the sensitivity of the detection method. Myhrvold et al. combined HUDSON technology with the SHERLOCK system to develop a highly sensitive and highly specific assay for dengue in body fluids (including urine, blood, saliva, serum and plasma). The virus and Zika virus are tested and the test results are given within 1 to 2 hours. Moreover, the preparation of the sample is also very simple, and does not require complicated equipment. In many parts of Central and South America, both dengue and Zika virus are very common, and clinical performance is similar. If pregnant women are infected with Zika virus, it will be very easy to produce congenital malformed children, and the virus can spread sexually, so it is especially important to test pregnant women and their husbands. This HUDSON-SHERLOCK detection technology can distinguish between dengue virus, Zika virus, and yellow fever virus (YFV), which are popular in many places in Brazil and cause serious epidemics. The technology also distinguishes four different subtypes of dengue virus, and can also perform single-nucleotide polymorphisms (SNP) analysis on different Zika virus. This function can be used for a variety of SNP analyses, such as resistance-related SNPs, virulence-related SNPs, and infectious related SNPs, to help us discover and track emerging pathogens. Myhrvold et al. also transformed the fluorescent signal of the SHERLOCK reporting system into a reporting system similar to the early pregnancy test strip, which is well suited for point-of care use.
The Gootenberg team also used the SHERLOCK technology, but they improved it and developed the SHERLOCKv2 detection system. This system allows simultaneous detection of three ssRNA targets and one dsDNA target in a single reaction. They biochemically tested 17 CRISPR-Cas13a enzymes and CRISPR-Cas13b enzymes, and then selected three enzymes with specific cleavage activity, supplemented by Cas12a enzyme and RPA system, and finally developed this ability in 90 minutes. Simultaneous detection of Zika virus ssRNA, synthetic ssRNA (synthetic ssRNA), dengue virus ssRNA, and synthetic dsDNA detection system, and the test results are still easy to interpret the visualization results. This SHERLOCKv2 technology, which detects both RNA and DNA, is ideal for the identification of pneumonia-causing pathogens. We know that pneumonia is the leading cause of death in young children around the world. Whether it is a DNA virus or RNA virus infection, or a combination of pneumonia caused by bacterial infection, it can be detected by this method. This convenient, inexpensive and convenient on-site pneumonia diagnosis technology can help patients with pneumonia in remote areas get early (antibacterial or antiviral) medical assistance.
Another SHERLOCKv2 technology enables highly sensitive quantitative detection of targets. This simple and accurate detection technology can monitor the HIV virus titer of AIDS patients in areas with relatively poor medical resources to judge the efficacy of antiviral therapy, thus improving the diagnosis and treatment of AIDS patients. This SHERLOCKv2 technology can also be used to detect mutations in the DNA molecules in the blood of patients with non-small cell lung cancer. The results can be either fluorescence readings or lateral flow assays, so they can also be used. For liquid biopsy. In the in vitro proof-of-concept test, the Gootenberg team also successfully applied SHERLOCKv2 technology for gene editing therapy, corrected colon cancer mutated genes, and also used SHERLOCKv2 technology to determine the number of successfully repaired mutated genes.
These newly developed diagnostic techniques also need to be compared to traditional detection techniques to ensure their sensitivity and specificity, as well as field testing to see if clinical testing needs are met, as different testing environments and different operators It may affect the test results. Once these tests are passed, these new detection technologies can be put into practical use to improve the level of diagnosis and treatment in remote areas. In areas with poor medical conditions, the diagnosis and treatment of unexplained fever is the most common and the most difficult to treat. These diseases are also diseases with high mortality and wide spread. For example, every year, 1.3 million people die from tuberculosis infection worldwide, so tuberculosis is also the disease with the highest mortality rate, and early detection and early treatment can actually prevent these patients from dying.
These detection techniques can be used to study the drug resistance of pathogenic pathogens and guide clinical use of drugs; the viability of pathogenic pathogens can also be analyzed to guide the prevention and control of infectious diseases; and the scope of detection can be extended to allow feces, Specimens such as respiratory secretions and cerebrospinal fluid are used as test samples to identify the causes of diseases such as enteritis, pneumonia, and meningitis. With the development of technology, according to the accuracy, reliability, simplicity, detection speed, flexibility, cost and other considerations, these detection techniques can also be applied to the detection of infectious diseases and non-communicable diseases, and can also be applied. In clinical, laboratory, and field applications.
Original search:
Daniel S. Chertow. (2018) Next-generation diagnostics with CRISPR. Science, 360: 381-382.
Source: Life Mystery / Eason
Tetanus Shot,Tetanus Vaccine,Hepatitis B Injection,Hep B Vaccine
FOSHAN PHARMA CO., LTD. , https://www.forepharm.com